Scaling of Polymer Solutions as a Quantitative Tool
Abstract
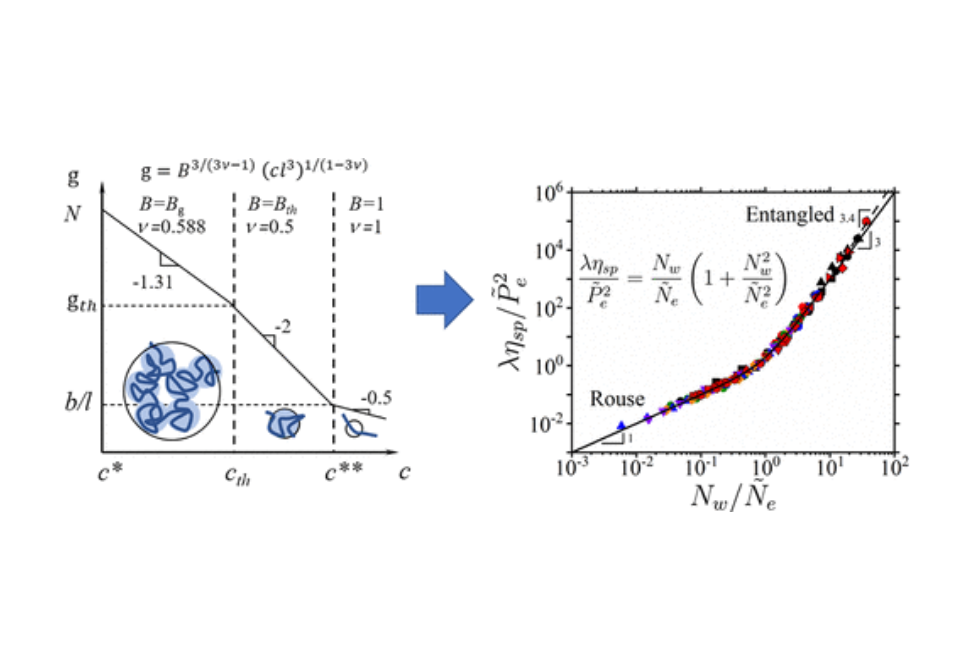
Knowledge of interaction parameters and Kuhn length for a polymer/solvent pair is a foundation of polymer physics of synthetic and biological macromolecules. Here, we demonstrate how to obtain these parameters from the concentration dependence of solution viscosity. The centerpiece of this approach is the scaling relationship between solution correlation length (blob size) ξ = lgν/B and the number of monomers per correlation blob g for polymers with monomer projection length l. The values of parameter B and exponent v are determined by solvent quality for the polymer backbone, chain Kuhn length, and types and strength of monomer–monomer and monomer–solvent interactions. Parameter B assumes values Bg, Bth, and 1 for exponent v = 0.588, 0.5, and 1, respectively. In particular, we take advantage of the linear relationship between specific viscosity ηsp in the unentangled (Rouse) regime and the number of correlation blobs Nw/g per chain with the weight average degree of polymerization, Nw, and g = B3/(3ν – 1)(cl3)1/(1 – 3ν) as a function of monomer concentration, c, and the corresponding B parameter. The values of the B parameters are extracted from the plateaus of normalized specific viscosity ηsp(c)/Nw(cl3)1/(3ν – 1) or their locations as a function of the monomer concentration c in different solution regimes. The extension of the approach to entangled polymer solutions provides a means to obtain the chain packing number, Pe, and to complete the set of parameters {Bg, Bth, Pe} (a system “fingerprint”) uniquely describing static and dynamic solution properties of a polymer/solvent pair. This approach is illustrated for solutions of poly(ethylene oxide) in water, poly(styrene) in tetrahydrofuran and toluene, poly(methyl methacrylate) in ionic liquids, and sodium carboxymethylcellulose in water at high salt concentrations.
Citation
Scaling of Polymer Solutions as a Quantitative Tool
Andrey V. Dobrynin, Michael Jacobs, and Ryan Sayko
Macromolecules 2021 54 (5), 2288-2295
DOI: 10.1021/acs.macromol.0c02810