RNA-Ligand Interactions Quantified by Surface Plasmon Resonance with Reference Subtraction
Abstract
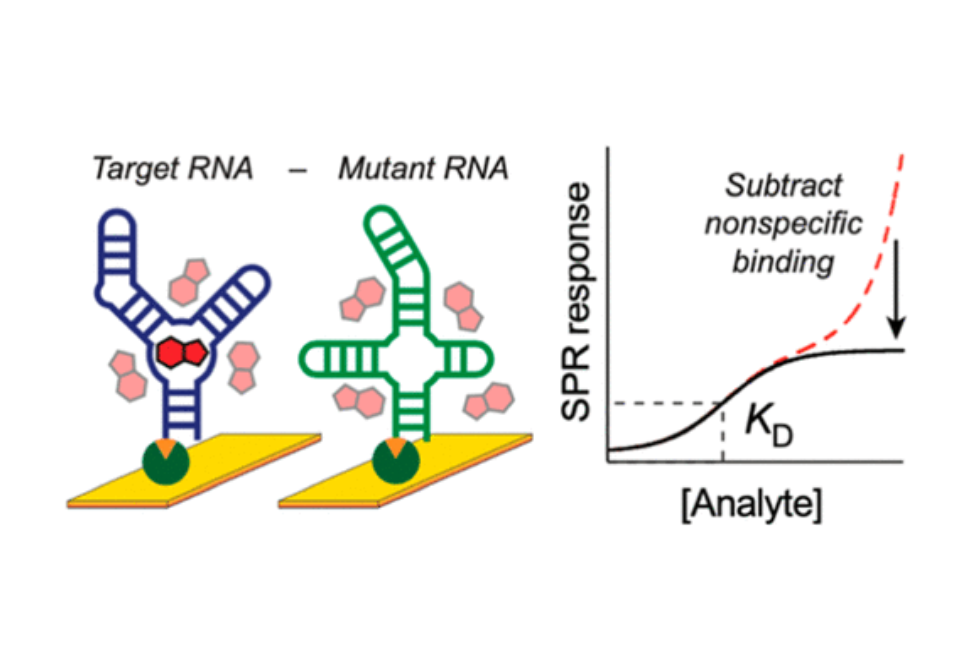
Structured RNAs bind ligands and are attractive targets for small-molecule drugs. A wide variety of analytical methods have been used to characterize RNA–ligand interactions, but our experience is that most have significant limitations in terms of material requirements and applicability to complex RNAs. Surface plasmon resonance (SPR) potentially overcomes these limitations, but we find that the standard experimental framework measures notable nonspecific electrostatic-mediated interactions, frustrating analysis of weak RNA binders. SPR measurements are typically quantified relative to a non-target reference channel. Here, we show that referencing to a channel containing a non-binding control RNA enables subtraction of nonspecific binding contributions, allowing measurements of accurate and specific binding affinities. We validated this approach for small-molecule binders of two riboswitch RNAs with affinities ranging from nanomolar to millimolar, including low-molecular-mass fragment ligands. SPR implemented with reference subtraction reliably discriminates specific from nonspecific binding, uses RNA and ligand material efficiently, and enables rapid exploration of the ligand-binding landscape for RNA targets.
Citation
RNA–Ligand Interactions Quantified by Surface Plasmon Resonance with Reference Subtraction
DOI: 10.1021/acs.biochem.2c00177