Double-stranded RNA drives SARS-CoV-2 nucleocapsid protein to undergo phase separation at specific temperatures
Abstract
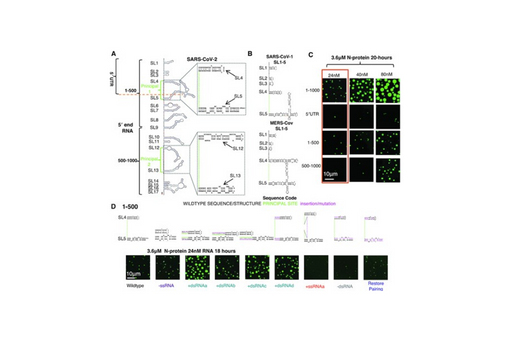
Nucleocapsid protein (N-protein) is required for multiple steps in betacoronaviruses replication. SARS-CoV-2-N-protein condenses with specific viral RNAs at particular temperatures making it a powerful model for deciphering RNA sequence specificity in condensates. We identify two separate and distinct double-stranded, RNA motifs (dsRNA stickers) that promote N-protein condensation. These dsRNA stickers are separately recognized by N-protein's two RNA binding domains (RBDs). RBD1 prefers structured RNA with sequences like the transcription-regulatory sequence (TRS). RBD2 prefers long stretches of dsRNA, independent of sequence. Thus, the two N-protein RBDs interact with distinct dsRNA stickers, and these interactions impart specific droplet physical properties that could support varied viral functions. Specifically, we find that addition of dsRNA lowers the condensation temperature dependent on RBD2 interactions and tunes translational repression. In contrast RBD1 sites are sequences critical for sub-genomic (sg) RNA generation and promote gRNA compression. The density of RBD1 binding motifs in proximity to TRS-L/B sequences is associated with levels of sub-genomic RNA generation. The switch to packaging is likely mediated by RBD1 interactions which generate particles that recapitulate the packaging unit of the virion. Thus, SARS-CoV-2 can achieve biochemical complexity, performing multiple functions in the same cytoplasm, with minimal protein components based on utilizing multiple distinct RNA motifs that control N-protein interactions.
Citation
Roden, C. A., Dai, Y., Giannetti, C. A., Seim, I., Lee, M., Sealfon, R., McLaughlin, G. A., Boerneke, M. A., Iserman, C., Wey, S. A., Ekena, J. L., Troyanskaya, O. G., Weeks, K. M., You, L., Chilkoti, A., & Gladfelter, A. S. (2022). Double-stranded RNA drives SARS-COV-2 nucleocapsid protein to undergo phase separation at specific temperatures. Nucleic Acids Research, 50(14), 8168–8192. https://doi.org/10.1093/nar/gkac596