Direct Mapping of Higher-Order RNA Interactions by SHAPE-JuMP
Abstract
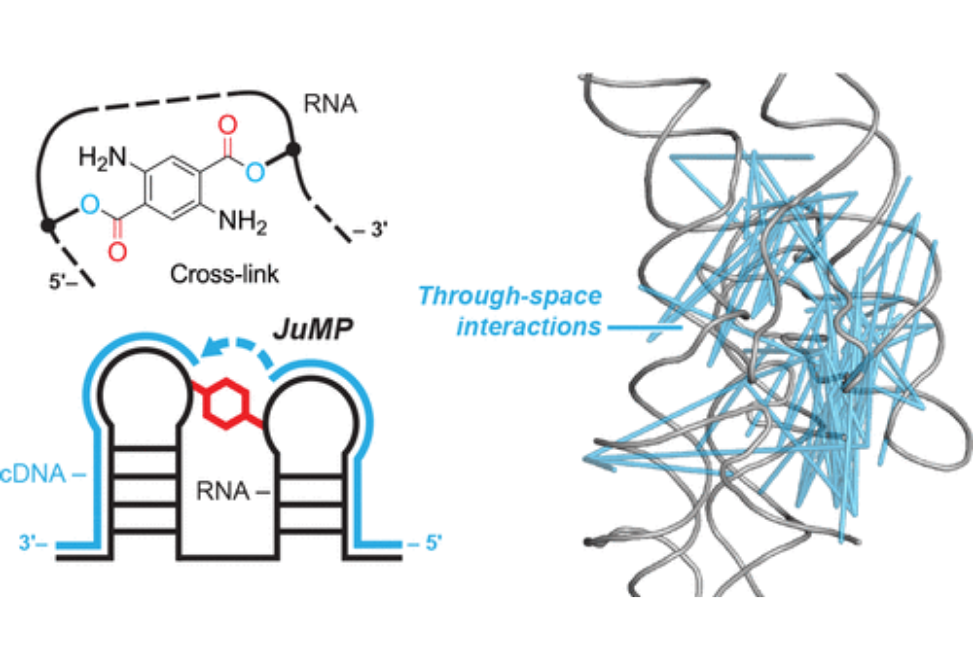
Higher-order structure governs function for many RNAs. However, discerning this structure for large RNA molecules in solution is an unresolved challenge. Here, we present SHAPE-JuMP (selective 2′-hydroxyl acylation analyzed by primer extension and juxtaposed merged pairs) to interrogate through-space RNA tertiary interactions. A bifunctional small molecule is used to chemically link proximal nucleotides in an RNA structure. The RNA cross-link site is then encoded into complementary DNA (cDNA) in a single, direct step using an engineered reverse transcriptase that “jumps” across cross-linked nucleotides. The resulting cDNAs contain a deletion relative to the native RNA sequence, which can be detected by sequencing, that indicates the sites of cross-linked nucleotides. SHAPE-JuMP measures RNA tertiary structure proximity concisely across large RNA molecules at nanometer resolution. SHAPE-JuMP is especially effective at measuring interactions in multihelix junctions and loop-to-helix packing, enables modeling of the global fold for RNAs up to several hundred nucleotides in length, facilitates ranking of structural models by consistency with through-space restraints, and is poised to enable solution-phase structural interrogation and modeling of complex RNAs.
Citation
Direct Mapping of Higher-Order RNA Interactions by SHAPE-JuMP
Thomas W. Christy, Catherine A. Giannetti, Gillian Houlihan, Matthew J. Smola, Greggory M. Rice, Jian Wang, Nikolay V. Dokholyan, Alain Laederach, Philipp Holliger, and Kevin M. Weeks
Biochemistry 2021 60 (25), 1971-1982
DOI: 10.1021/acs.biochem.1c00270