Enzymatic Macrolactamization of mRNA Display Libraries for Inhibitor Selection
Abstract
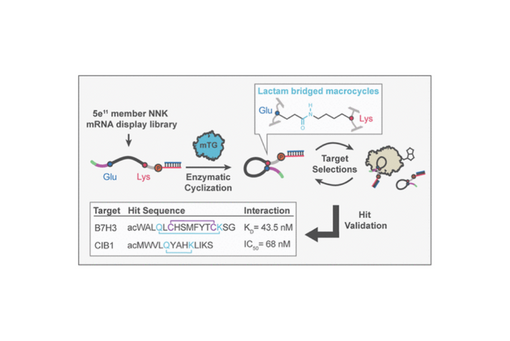
mRNA display is a powerful, high-throughput technology for discovering novel, peptide ligands for protein targets. A number of methods have been used to expand the chemical diversity of mRNA display libraries beyond the 20 canonical amino acids, including genetic code reprogramming and biorthogonal chemistries. To date, however, there have been few reports using enzymes as biocompatible reagents for diversifying mRNA display libraries. Here, we report the evaluation and implementation of the common industrial enzyme, microbial transglutaminase (mTG), as a versatile biocatalyst for cyclization of mRNA display peptide libraries via lysine-to-glutamine isopeptide bonds. We establish two separate display-based assays to validate the compatibility of mTG with mRNA-linked peptide substrates. These assays indicate that mTG has a high degree of substrate tolerance and low single round bias. To demonstrate the potential benefits of mTG-mediated cyclization in ligand discovery, high diversity mTG-modified libraries were employed in two separate affinity selections: (1) one against the calcium and integrin binding protein, CIB1, and (2) the second against the immune checkpoint protein and emerging therapeutic target, B7–H3. Both selections resulted in the identification of potent, cyclic, low nanomolar binders, and subsequent structure–activity studies demonstrate the importance of the cyclization to the observed activity. Notably, cyclization in the CIB1 binder stabilizes an α-helical conformation, while the B7–H3 inhibitor employs two bridges, one mTG-derived lactam and a second disulfide to achieve its potency. Together, these results demonstrate potential benefits of enzyme-based biocatalysts in mRNA display ligand selections and establish a framework for employing mTG in mRNA display.
Citation
Enzymatic Macrolactamization of mRNA Display Libraries for Inhibitor Selection
Matthew M. Bowler, Marta Glavatskikh, Chad V. Pecot, Dmitri Kireev, and Albert A. Bowers
ACS Chemical Biology 2023 18 (1), 166-175
DOI: 10.1021/acschembio.2c00828