Subsite Ligand Recognition and Cooperativity in the TPP Riboswitch: Implications for Fragment-Linking in RNA Ligand Discovery
Abstract
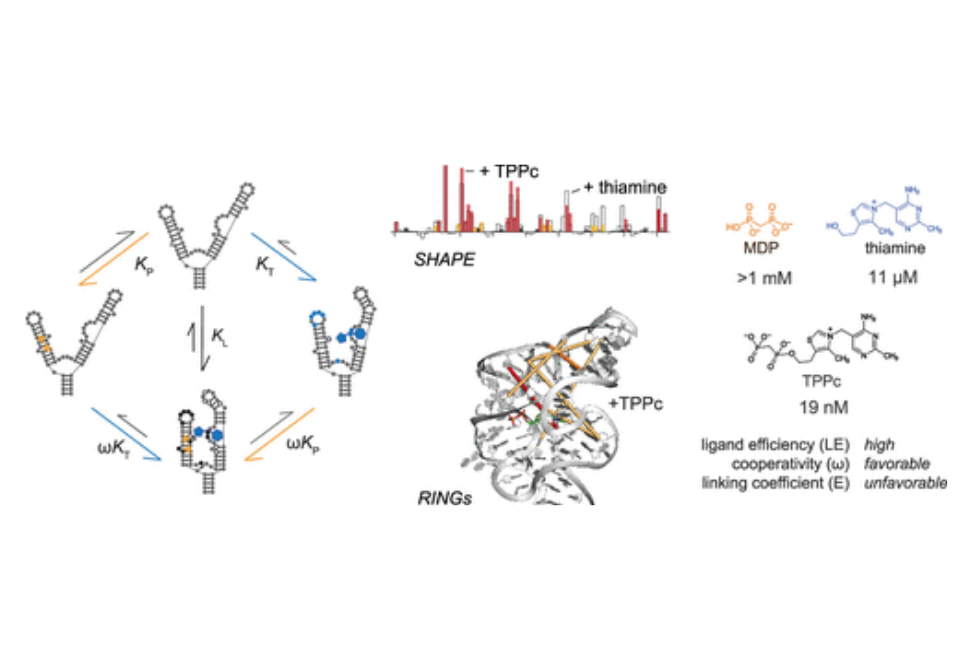
RNA molecules can show high levels of cooperativity in their global folding and interactions with divalent ions. However, cooperativity at individual ligand–RNA interaction sites remains poorly understood. Here, we investigated the binding of thiamine and methylene diphosphonic acid (MDP, a soluble structural analogue of pyrophosphate) to the thiamine pyrophosphate riboswitch. These ligands each bind weakly at proximal subsites, with 10 μM and 1 mM affinities, respectively. The affinity of MDP moderately improves when thiamine or thiamine-like fragments are pre-bound to the RNA. Covalent linking of thiamine and MDP substantially increases riboswitch binding to a notable high affinity of 20 nM. Crystal structures and single-molecule correlated chemical probing revealed favorable induced fit effects upon binding of individual ligands and, unexpectedly, a substantial thermodynamically unfavorable RNA structural rearrangement upon binding of the linked thiamine–MDP ligand. Thus, linking of two ligands of modest affinity, accompanied by an unfavorable structural rearrangement, still yields a potent linked RNA-binding compound. Since complex ligands often bind riboswitches and other RNAs at proximal subsites, principles derived from this work inform and support fragment-linking strategies for identifying small molecules that interact with RNA specifically and with high affinity.
Citation
Subsite Ligand Recognition and Cooperativity in the TPP Riboswitch: Implications for Fragment-Linking in RNA Ligand Discovery
Meredith J. Zeller, Ashok Nuthanakanti, Kelin Li, Jeffrey Aubé, Alexander Serganov, and Kevin M. Weeks
ACS Chemical Biology Article ASAP
DOI: 10.1021/acschembio.1c00880