Faculty Members

Kevin Weeks
Kenan Distinguished Professor
Genome Sciences Building 3258919-962-7486
weeks@unc.edu
Group Website
Curriculum Vitae
Research Interests
Chemical Biology, RNA Structure & Function, RNA Therapeutics
Research Synopsis
Research in the Weeks group lies at the interface of chemistry, biology and genomics. One of the most amazing discoveries of recent years has been the profound role of RNA in regulating all areas of biology. For example, a much larger fraction of the human genome is transcribed into RNA than codes for protein synthesis. Further, the functions of many RNA molecules require that an RNA fold back on itself to create intricately and complexly folded structures. Until recently, however, we had little idea of the broad contributions of RNA structure and function because there simply did not exist rigorous chemical tools for understanding RNA molecules in cells and viruses.
The vision of our laboratory is therefore, first, to invent novel chemical microscopes that reveal quantitative structure and function interrelationships for RNA and, second, to apply these RNA technologies to broadly important problems in biology.
Our work is highly interdisciplinary. Projects in the laboratory meld fundamental basic science chemistry, computational chemistry, and biotechnology development and extend to practical applications in virology, high-throughput RNA structure analysis, and understanding biological processes in cells.
Current projects focus on (i) RNA folding and protein assembly reactions central to the replication of human viruses, including HIV and Dengue, (ii) function of biomedically important RNA-protein complexes inside living cells, and (iii) discovery of small molecule ligands, potential drugs, targeted against medically important RNAs.
Professional Background
Fulbright Scholar, Universität Göttingen, Germany, 1987; Yale University, Ph.D., 1992; Jane Coffin Childs Postdoctoral Fellow, University of Colorado, 1992-1996; Research Innovation Award, Research Corporation, 1997; Searle Scholar in the Biomedical Sciences, 1998-2001; NSF Career Award, 2000-2005; Visiting Scholar, National Institutes of Health, NIEHS, 2002-2003; North Carolina Health & Life Sciences Promise for Tomorrow Award, 2009; NIH EUREKA Award, 2011-2015; Fellow, American Association for the Advancement of Science, 2012; Kenan Senior Faculty Competitive Research and Scholarly Leave, 2013; Life Member, Clare Hall, University of Cambridge, UK, 2014
News & Publications
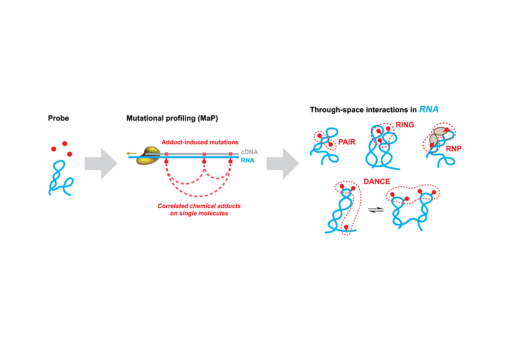
Enabled by a specialized reverse transcription strategy called mutational profiling (MaP), multiple chemical modification events can be measured within individual RNA molecules.
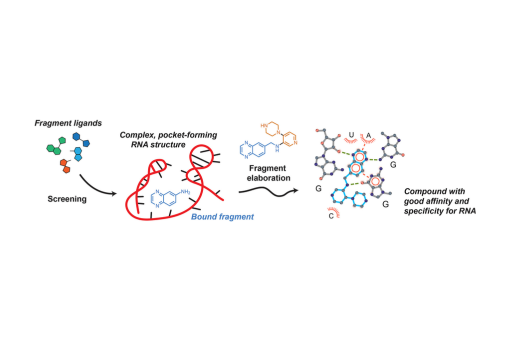
Here we share an integrated analysis of recent innovations in FBLD, emphasizing opportunities resulting from fragment elaboration via both linking and growing.

Scott Warren
Associate Professor
Kenan Laboratories A808919-966-0994
scw@email.unc.edu
Group Website
Curriculum Vitae
Research Interests
2D Materials, Energy Storage, Solar Energy, Nanoelectronics
Research Synopsis
2D materials are a radically new building block for constructing complex materials and devices, from batteries to solar cells to water purification to sensors. Our lab is focused on:
1. The discovery of new 2D materials
2. The assembly of these building blocks into functional architectures
3. Understanding structure-property relationships in 2D materials and their assemblies
4. Integrating these new materials into devices for energy conversion, environmental remediation, and sensing
Our lab is interdisciplinary and collaborative, and we draw on both experimental approaches, such as materials synthesis, electrochemistry, electronics, nanophotonics, and electron microscopy, and computational approaches, such as density functional theory, finite difference time domain simulations, and multislice calculations, to understand the materials that we make.
Professional Background
Research Group
News & Publications

Enabled by a specialized reverse transcription strategy called mutational profiling (MaP), multiple chemical modification events can be measured within individual RNA molecules.

Here we share an integrated analysis of recent innovations in FBLD, emphasizing opportunities resulting from fragment elaboration via both linking and growing.

Domenic Tiani
Teaching Professor
Kenan Laboratories B128919-962-1616
tiani@email.unc.edu
Curriculum Vitae
Research Interests
Chemistry Education, Lab Curriculum Development
Research Synopsis
My primary research interests lie in the area of chemical education. In particular, I am interested in the development and implementation of new and better methods by which to teach fundamental chemical concepts in the classroom and laboratory. As a chemical educator my objectives are to: Provide a challenging and stimulating learning environment, one in which students are active participants in the learning process. Provide connectivity between what they learn in the classroom, what they do in the laboratory and what they see in the world around them. Train students to think critically and be problem solvers. Develop research projects focused on investigating fundamentally important and interesting chemical problems, and involve undergraduate students in these projects. Currently my role in the undergraduate chemistry program at UNC-CH involves undergraduate instruction, curriculum development and the training/supervision of graduate students as laboratory teaching assistants. With regards to curriculum development, I am involved with the "Separation and Analytical Characterization of Organic and Biological Compounds" laboratory course. My role in this course is three-fold. First, I ensure that the laboratory manual/material is current and is a useful text/resource for those students taking the course. Secondly, I work to develop experiments for this course that are relevant and in-line with the course objectives. Finally, I work to make sure that the laboratory resources, such as computers, software and instrumentation, remain current so that students continue to learn a variety of modern experimental techniques. All of the above objectives are each important in guaranteeing that a student's learning experience at UNC-CH is both meaningful and rewarding.
Professional Background
North Carolina State University, B. A., 1995; Phi Beta Kappa, NCSU, 1995; Phi Kappa Phi, NCSU, 1995; Laboratory Technician, LabCorp, 1996-1997; Laboratorian of the Year, LabCorp, 1997; University of Arizona, Ph.D., 1997-2003; Graduate Teaching Award, University of Arizona, 1998; Lecturer Adjunct Professor in Chemistry, University of Arizona, 2002-2003; Tanner Award for Excellence in Undergraduate Teaching, 2016
News & Publications

Enabled by a specialized reverse transcription strategy called mutational profiling (MaP), multiple chemical modification events can be measured within individual RNA molecules.

Here we share an integrated analysis of recent innovations in FBLD, emphasizing opportunities resulting from fragment elaboration via both linking and growing.

Joseph Templeton
Francis Preston Venable Professor of Chemistry
Kenan Laboratories 448919-966-4575
joetemp@unc.edu
Curriculum Vitae
Research Interests
Organometallic Chemistry
Research Synopsis
Organometallic projects currently underway in our laboratory involve bond activation with platinum and palladium, tungsten carbene and carbyne chemistry, and stereoselective addition reactions of coordinated ligands. Three specific projects are summarized below.
The utility of transition metal carbene reagents has blossomed dramatically during the past decade. Electron-rich carbene complexes, [Tp'(OC)2W=CHR]- anions have been prepared, and these complexes exhibit unusual reactivity patterns at the carbene carbon. The availability of a heteroatom carbene complex, Tp'I(O)W=C(CH3)(OSiR3), in the Schrock manifold of oxidation states provides an opportunity to define the reactivity patterns available to this class of carbene complexes.
We have developed robust Pt(IV) organometallic complexes containing alkyl, hydride, aryl, acyl, formyl and silyl ligands in various combinations with Tp' anchoring the octahedral coordination sphere. The relevance of Pt(II)/Pt(IV) interconversions to alkane activation processes provide the driving force for these synthetic efforts.
The chiral [Tp'W(CO)(PhCºCR)]+ fragment binds ketones and imines. These coordinated ketones or imines readily add nucleophiles at carbon, and we are exploring the stereochemistry of the reduced products.
Professional Background
Iowa State University, Ph.D., 1975; Postdoctoral research as NATO fellow at Imperial College of Science and Technology, 1975-1976; Fellow, American Association for the Advancement of Science.
News & Publications

Enabled by a specialized reverse transcription strategy called mutational profiling (MaP), multiple chemical modification events can be measured within individual RNA molecules.

Here we share an integrated analysis of recent innovations in FBLD, emphasizing opportunities resulting from fragment elaboration via both linking and growing.

Sergei Sheiko
George A. Bush, Jr. Distinguished Professor
Caudill Laboratories 157919-843-5270
sergei@email.unc.edu
Group Website
Curriculum Vitae
Research Interests
Tissue-Adaptive Materials, Programmable Materials Design, and Reconfigurable Polymer Networks
Research Synopsis
Materials science strives for intelligent soft materials that are able to sense, process, and adapt to their environment. Applications range from tissue engineering and microelectronics to renewable energy and climate change.
Within this broad area of research, we are particularly interested in the programmable design of tissue-mimetic materials for biomedical devices, soft robotics, and wearable electronics. We want to develop macromolecules that self-assemble to elastomers and gels that precisely replicate the mechanics of living tissues ranging from brain to skin. Unlike conventional trial-and-error experimentation with chemical formulations, our approach is based on encoding materials properties in molecular architecture. In line with artificial intelligence, the design-by-architecture approach enables the efficient synthesis of well-defined materials with predictable properties that can be adjusted on demand for customizable applications, particularly personalized medicine. We are currently developing chemistry for minimally invasive injection and insertion of non-leaching implants to the human body followed by an adaptive matching to the surrounding tissue mechanics. This platform can be readily extended to 3D printing, injection molding of biomedical devices, and waste-free plastics upcycling.
Professional Background
Fellow, American Physical Society, 2010, Professor, University of North Carolina at Chapel Hill, 2001-present, Habilitation - University of Ulm, Germany, 2001, Postdoctoral Fellow - University of Twente, The Netherlands, 1991-1993, PhD - Institute of Chemical Physics of the Russian Academy of Sciences, 1991, BS - Moscow Physico-Technical Institute, 1986.
Research Group
News & Publications

Enabled by a specialized reverse transcription strategy called mutational profiling (MaP), multiple chemical modification events can be measured within individual RNA molecules.

Here we share an integrated analysis of recent innovations in FBLD, emphasizing opportunities resulting from fragment elaboration via both linking and growing.

Mark Schoenfisch
Peter A. Ornstein Distinguished Professor, Jointly appointed with the School of Medicine & School of Pharmacy
Caudill Laboratories 336919-843-8714
schoenfisch@unc.edu
Group Website
Curriculum Vitae
Research Interests
Analytical Sensors, Biomaterials & Nanoparticle Therapeutics
Research Synopsis
Our research is focused on four main areas: 1) designing macromolecular nitric oxide release vehicles as novel therapeutics; 2) improving the analytical performance of implantable continuous glucose monitoring devices for diabetes management; 3) designing microfluidic nitric oxide, NO, sensors for real-time detection of NO in biological media; and, 4) developing superhydrophobic interfaces for mold prevention and remediation.
In the Schoenfisch Lab, we work at the interface of analytical chemistry, materials science, biomedical engineering, and biology. The types of multi-disciplinary research opportunities that are available include studies of therapeutics to treat various diseases; sensors that function reliably and continuously, real time, to facilitate disease management; microelectrode and microfluidic sensor design and fabrication for clinical, point-of-care, and diagnostic/prognostic applications; and, new macromolecular scaffolds that manipulate biology and physiology.
Professional Background
University of Kansas, B.A., 1992; University of Arizona, Ph.D., 1997; University of Michigan, National Institutes of Health Postdoctoral Fellow, 1998-1999; Society for Analytical Chemists of Pittsburgh Young Investigator Award, 2001; Eli Lilly and Company Young Investigator Award, 2002-2004; National Science Foundation CAREER Award, 2004-2009; International Union of Pure and Applied Chemistry Young Observer Award , 2005; John L. Sanders Award for Excellence in Undergraduate Teaching and Service, 2007; Chapman Family Teaching Award for Distinguished Teaching of Undergraduate Students, 2015
News & Publications

Enabled by a specialized reverse transcription strategy called mutational profiling (MaP), multiple chemical modification events can be measured within individual RNA molecules.

Here we share an integrated analysis of recent innovations in FBLD, emphasizing opportunities resulting from fragment elaboration via both linking and growing.

Matthew Redinbo
Kenan Distinguished Professor of Chemistry, Biochemistry and Microbiology, Jointly appointed with the School of Medicine
Genome Sciences Building 4350919-962-4581
redinbo@unc.edu
Group Website
Curriculum Vitae
Research Interests
Understanding Human Disease using Structural and Chemical Biology
Research Synopsis
Inhibitors Intercept the Catalytic Cycle of Gut Microbial Enzymes
In research published in ACS Central Science, researchers led by Matthew Redinbo, and in work largely conducted by graduate student Samuel Pellock, demonstrate that unique chemotypes can inhibit gut microbial b-glucuronidase, GUS, enzymes by high-jacking the catalytic cycle of these well-studied proteins.
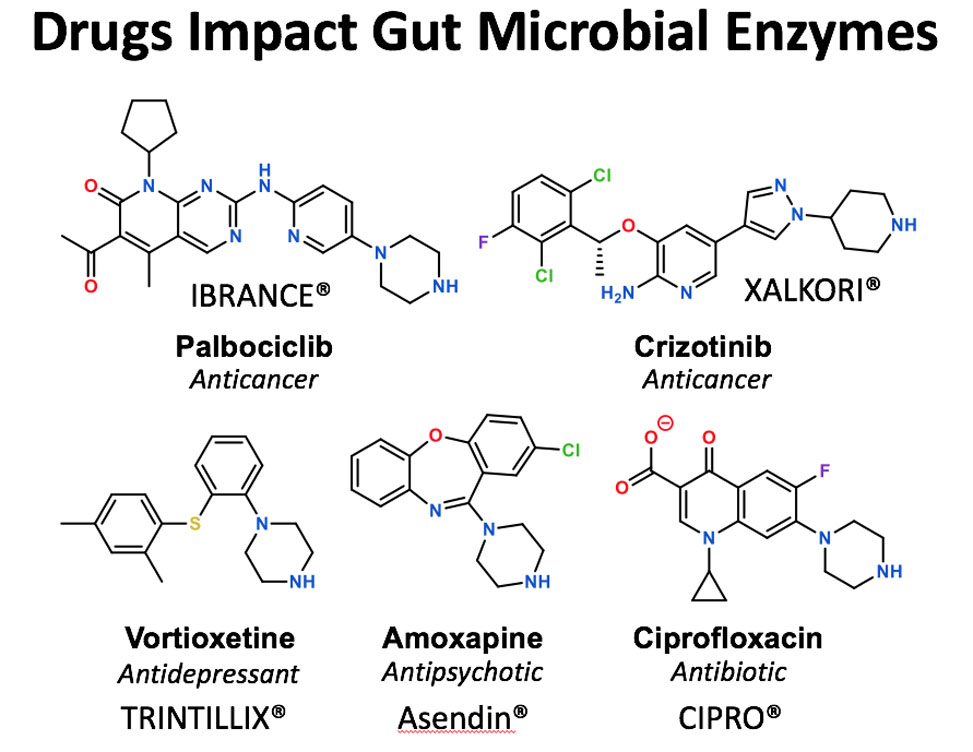
GUS enzymes are members of the glycoside hydrolase family of enzymes, some of the first examined in detail. Yet, in spite of decades of effort, the work presented by Pellock and colleagues shows that distinct, piperizine-containing compounds, including many current drugs, can potently inhibit the reactions carried out by these enzymes by replacing water in the second half of their catalytic cycle. The research suggests that GUS enzymes in the gut can be potently controlled using these reagents.
Gut microbial GUS proteins are important for the efficacy and tolerance of a wide range of drugs, which reach the gut as drug-glucuronides and can serve as GUS substrates. Blocking the reactivation of drugs in the gut using such reagents has been shown by Redinbo and collaborators to block the toxicity of cancer drugs and NSAIDs. This work further shows that existing drugs are impacting the gut microbiota in previously unknown ways, and with consequences that have yet to be appreciated.
Transcriptional Regulation in Gut Microbial Pathogens
Work published in PNAS by graduate student Michael Little, and led by Professor Matthew Redinbo, has shown for the first time how a group of gut microbial pathogens control the expression of a key operon of factors when a certain source of energy is available. They have focused on the Enterobacteriaceae, a family of opportunistic gut bacterial pathogens that include microbes like Salmonella, Shigella, Yersinia and Escherichia coli. These bacteria can grow up in the damaged GI tract and lead to local or systemic infections, and they are unique in the gut microbial world in that they encode their b-glucuronidase, GUS, enzyme and related glucuronide scavenging machinery on a operon controlled by a single transcription factor, GusR.
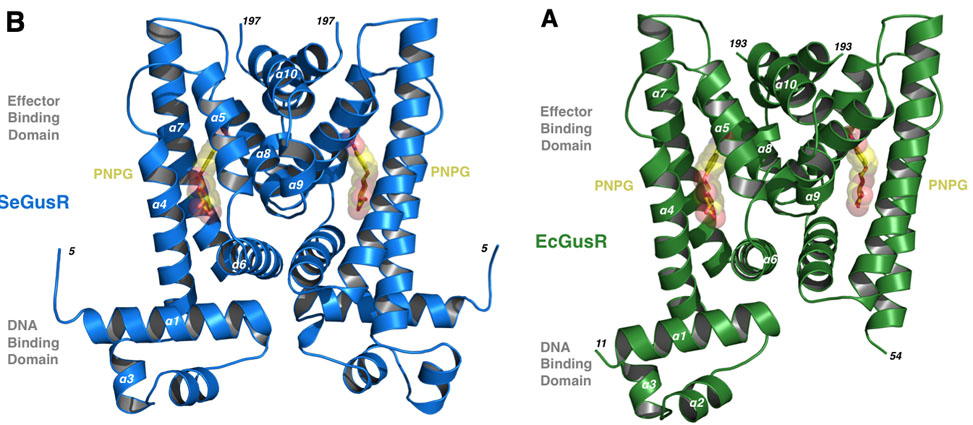
In work by Little and colleagues, the structures of GusR proteins from Salmonella and E. coli are presented, and they are shown to respond to glucuronide conjugates found in the human GI tract, and in response to these compound they up-regulate the expression of GUS activity. This work shows that these relatively trace organisms in the complex gut microbial milieu have a "turbo-charger" to express GUS to get the glucuronide sugar as an energy source, and suggest that these microbes may be reliant on GusR and GUS during infection, which could lead to new therapeutics to block subsequent infections.
Discovery Blocks Cancer Drug's Toxic Side Effect
Findings published in Science from the Redinbo Group, in collaboration with UNC School of Medicine, the Albert Einstein College of Medicine, and North Carolina Central University, may lead to the elimination of a debilitating side effect of CPT-11, a widely used but harshly potent treatment for colon cancer.
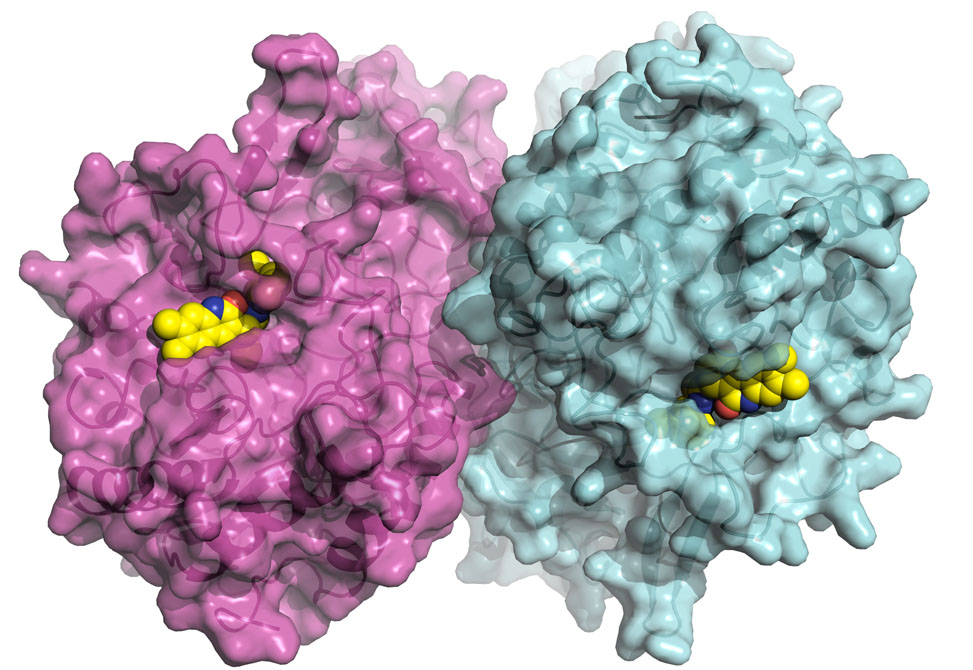
The team of researchers, led by chemistry professor Matthew Redinbo from the University of North Carolina at Chapel Hill, has discovered that it is possible to target and block the enzyme, beta glucuronidase, which is thought to play a major role in causing the drug's side effects. "In a manner of speaking, we cured the bacteria's sweet tooth without damaging the microbes or intestines and, in the process, the drug's toxic side effect was alleviated," said Redinbo.
Study co-author, Sridhar Mani, professor of medicine and genetics at Einstein, said the severe diarrhea caused by CPT-11 can sharply limit the dosage that cancer patients can receive. "Our tests showed conclusively that the inhibitor identified by our UNC colleagues prevented diarrhea in mice that were also receiving CPT-11. We are hopeful that clinical trials will show that administering this inhibitor when patients start taking CPT-11 allows for improvement in the drug's anti-tumor effect in patients with cancer."
Listen to "The Story" interview with Dick Gordon on WUNC, November 9, 2010
Professional Background
B.S., 1990, University of California, Davis; Ph.D., 1995, University of California, Los Angeles; Postdoctoral Fellow, 1995-1999, University of Washington, Seattle; Visiting Professor, 2013-2014, SGC, Nuffield Department of Medicine; and Visiting Fellow, Magdalen College; Oxford University; Outstanding Mentor Award, 2018; Fellow, AAAS, 2013; Academic Leadership Fellow, UNC, 2011; Phillip and Ruth Hettleman Prize for Artistic and Scholarly Achievement, 2004; Burroughs Wellcome Career Award in the Biomedical Sciences 1999


Research Group
Researchers in the Redinbo Laboratory use the tools of structural, molecular and chemical biology to examine a range of dynamic cellular processes central to human health.
Current projects include the discovery of new antimicrobials targeted to drug-resistant bacteria, the design of novel proteins engineered to detect and eliminate toxic chemicals, and the development of small-molecule to cell-based methods to improve anticancer chemotherapeutics.
We also continue our focus on determining the crystal structures of macromolecular complexes, including those involving human nuclear receptors central to transcriptional regulation, bacterial proteins involved in DNA manipulation and human cell contact, and enzymes central to key cellular processes.
News & Publications

Enabled by a specialized reverse transcription strategy called mutational profiling (MaP), multiple chemical modification events can be measured within individual RNA molecules.

Here we share an integrated analysis of recent innovations in FBLD, emphasizing opportunities resulting from fragment elaboration via both linking and growing.

Michael Ramsey
Minnie N. Goldby Distinguished Professor of Chemistry; Jointly appointed with the School of Medicine
Chapman Hall 251919-962-7492
jmramsey@unc.edu
Group Website
Curriculum Vitae
Research Interests
Microfabricated Chemical Instrumentation, Microfluidics, Nanofluidics
Research Synopsis
We are interested in utilizing micro- and nanofabrication strategies to create devices that facilitate our abilities to gather chemical and biochemical information. Our motivations for fabricating devices include point of care clinical diagnostics, high-throughput biochemical experimentation, development of new types of chemical sensors, and understanding of transport mechanisms in nanoscale-confined spaces. The devices that we develop have application to drug discovery, health care, environmental monitoring, and basic research.
Many of our projects involve the microfabrication of fluidic networks that are used to perform biochemical assays. This area of research is often referred to as microfluidics, microchips, or Lab-on-a-Chip technology. We are presently interested in developing microfluidic devices for addressing clinical diagnostics, proteomics problems, and automated single cell assays. Proteomics is a term used to describe the understanding of the full complement of proteins expressed by an organism. A given type of cell will contain several thousand proteins and the task of identifying all proteins is a significant challenge. Traditional approaches to solving this problem involve the use of milligram quantities of protein and chemical separations that unfold over a period of days. We are pursuing microfabricated fluidics approaches to implementing multidimensional liquid phase separations that utilize nanogram to picogram quantities of materials with separations unfolding over a period of tens of minutes. The ability to perform electrospray ionization from microchips is also being developed to allow structural analysis via mass spectrometry of the isolated proteins.
Microfluidics devices are also being developed to autonomously manipulate single non-adherent cells, characterize them using flow cytometry techniques, lyse the cells and chemical analyze the cell contents utilizing chemical separations techniques. This automated approach allows high-throughput characterization at the single cell level so that statistics can be rapidly generated on cell heterogeneity. Through collaboration with Prof. Nancy Allbritton, UNC-CH, multiple kinase assays are being implemented on these devices to study cell signal transduction mechanisms.
More fundamental in nature is the study of molecular transport through conduits with nanometer scale dimensions, what we refer to as nanofluidics. We are studying both fluid and polymer transport through individual nanoscale conduits or pores that are top-down fabricated in hard materials such as glass, quartz, or silicon. We are attempting to further reduce lateral dimensions of channels and pores utilizing bottom-up strategies and interfacing molecular assemblies to features formed in hard materials. While this work is fundamental, understanding could lead to new methods for the separation and analysis of polymeric materials, new types of chemical sensors, or possibly technology that allows the structure of single polymer molecules to be determined, for example, single molecule DNA sequencing.
We are also investigating the prospects of shrinking the size of conventional mass spectrometry by several orders of magnitude. Specifically we have been experimentally exploring the reduction in size of Paul-type ion trap mass analyzers and assessing their performance. Reduction in length scale of this type of mass spectrometer is particularly interesting because the mass resolution of the device does not fundamentally depend on its size. Microscale ion trap MS devices that are a factor of 40 smaller than conventional scale have been operated with no loss in mass resolving power. Microfabricated ion trap devices with fundamental length scales as small as 10 μm have been demonstrated. Smaller mass analyzers also allow operation at lower ion mean-free path, higher pressures. Mass spectra have been generated at pressures exceeding 1 torr, greatly reducing pumping requirements. Miniature mass spectrometers have application to problems such as environmental monitoring, chemical process control, and high-throughput laboratory analysis.
Professional Background
B.S. Chemistry, Bowling Green State University, 1974; Ph.D. Chemistry, Indiana University, 1979; Eugene P. Wigner Distinguished Postdoctoral Fellow, Oak Ridge National Laboratory, 1979; Research Staff, Oak Ridge National Laboratory, 1981; Group Leader, Oak Ridge National Laboratory, 1985; Alumnus of the Year, Department of Chemistry, Bowling Green State University, 1992; Founding Scientist, Caliper Technologies, Inc., 1995; Scientist of the Year Award, Oak Ridge National Laboratory, 1996; Discover Magazine Annual Technology Award, 1996; R&D 100 Award, Lab-on-a-Chip Technology, 1996; Corporate Research Fellow, Oak Ridge National Laboratory, 1997; Fellow, Optical Society of America, 1998; Alexander von Humboldt Award for Senior Scientist, 1999; Frederick Conference Capillary Electrophoresis Award, 2000; Desty Memorial Lecture, The Royal Institution, 2000; R&D 100 Top 40, Lab-on-a-Chip Technology, 2001; Energy@23 Award, 2001; A. J. P. Martin Gold Medal for Separation Science, 2001; Jacob Heskel Gabbay Award in Biotechnology and Medicine, 2001; Battelle Distinguished Inventor Award, 2003; Marcel J. E. Golay Award in Capillary Chromatography, 2003; ACS Division of Analytical Chemistry Award in Chemical Instrumentation, 2003; R&D 100 Award, μTrapMS, 2003; Federal Laboratory Consortium Excellence in Technology Transfer Award, 2004; Pittsburgh Analytical Chemistry Award, 2006; American Chemical Society Award in Chromatography, 2007; Fellow, American Institute for Medical and Biological Engineering, 2008, Fellow, American Chemical Society, 2011; The CASSS Award for Outstanding Achievements in Separation Science, 2012; Ralph N. Adams Award in Bioanalytical Chemistry, 2013; Member, National Academy of Engineering, 2014
Research Group
News & Publications

Enabled by a specialized reverse transcription strategy called mutational profiling (MaP), multiple chemical modification events can be measured within individual RNA molecules.

Here we share an integrated analysis of recent innovations in FBLD, emphasizing opportunities resulting from fragment elaboration via both linking and growing.

Gary Pielak
Kenan Distinguished Professor of Chemistry, Biochemistry and Biophysics; Jointly appointed with the School of Medicine
Genome Sciences Building 3250919-962-4495
gary_pielak@unc.edu
Group Website
Curriculum Vitae
Research Interests
Protein Biophysics Emphasizing Studies in Living Cells
Research Synopsis
My graduate students and I work in two areas: High-resolution protein NMR studies in living cells. Biophysics of tardigrade dessication-tolerance proteins.
Professional Background
BS Chem., Bradley U, 1977; PhD Biochemistry, with J. Ivan Legg, Washington State U, 1983; Postdoc with Michael Smith, U British Columbia, 1983-1986; Postdoc with Robert JP Williams, Oxford. 1986-1988; DuPont Young Faculty Award; Morrow Young Faculty Award; NIH Pioneer Award; National Science Foundation Program Director; UNC Lifetime Mentor Award; UNC Medical School Excellence in Basic Science Mentoring Award
Research Group
News & Publications

Enabled by a specialized reverse transcription strategy called mutational profiling (MaP), multiple chemical modification events can be measured within individual RNA molecules.

Here we share an integrated analysis of recent innovations in FBLD, emphasizing opportunities resulting from fragment elaboration via both linking and growing.

David Nicewicz
Kenan Distinguished Professor
Murray Hall 2202G919-843-9150
nicewicz@unc.edu
Group Website
Curriculum Vitae
Research Interests
Synthetic Organic Chemistry, Catalysis, Natural Product Synthesis
Research Synopsis
My research interests lie in the broadly defined fields of asymmetric catalysis and target-oriented synthesis. Concerning the area of asymmetric catalysis, my laboratory will focus primarily on the development of novel catalytic processes to access highly reactive intermediates under operationally mild conditions. Methods encompassing the general areas of electron transfer and atom abstraction are of particular interest. In the context of natural product synthesis, targets will be selected on the basis of their biological activity and structural complexity. Specifically, I am interested in complex molecules that inspire the development of new strategies and methods for their construction.
Professional Background
B.S., University of North Carolina at Charlotte, 2000; M.S., University of North Carolina at Charlotte, 2002; Ph.D., University of North Carolina at Chapel Hill, 2006; NIH Postdoctoral Fellow, Princeton University, 2007-2009; Eli Lilly New Faculty Award, 2009; Packard Fellowship in Science and Engineering, 2012; New Investigator Award in Organic Chemistry, sponsored by Boehringer Ingelheim, 2013; NSF CAREER Award, 2014-2019; Amgen Young Investigator Award, 2014
Research Group
News & Publications

Enabled by a specialized reverse transcription strategy called mutational profiling (MaP), multiple chemical modification events can be measured within individual RNA molecules.

Here we share an integrated analysis of recent innovations in FBLD, emphasizing opportunities resulting from fragment elaboration via both linking and growing.