Time-Resolved, Single-Molecule, Correlated Chemical Probing of RNA
Abstract
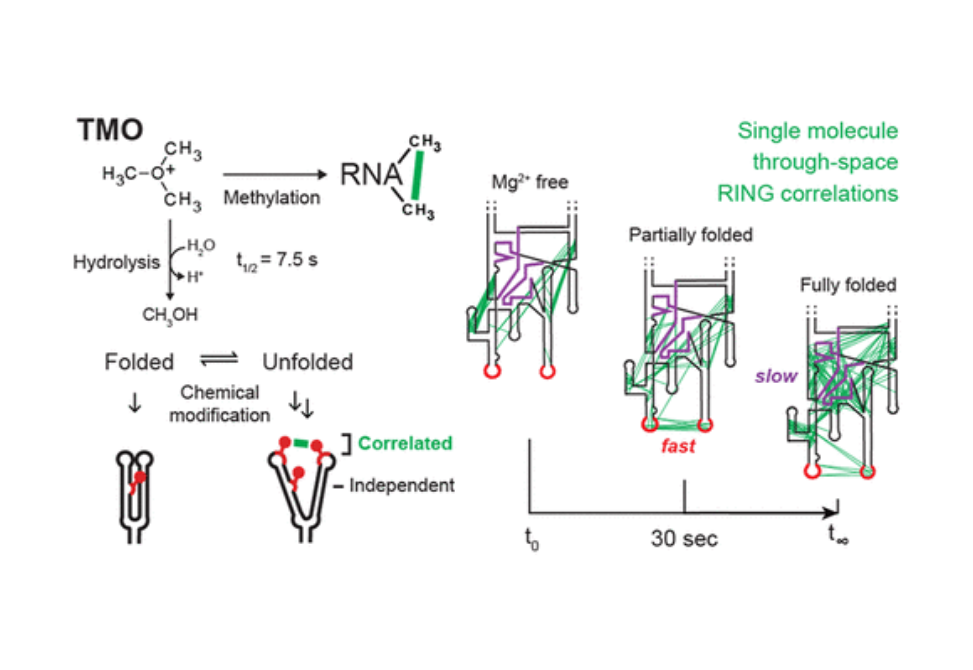
Capturing the folding dynamics of large, functionally important RNAs has relied primarily on global measurements of structure or on per-nucleotide chemical probing. These approaches infer, but do not directly measure, through-space structural interactions. Here we introduce trimethyloxonium (TMO) as a chemical probe for RNA. TMO alkylates RNA at high levels in seconds, and thereby enables time-resolved, single-molecule, through-space probing of RNA folding using the RING-MaP correlated chemical probing framework. Time-resolved correlations in the RNase P RNA—a functional RNA with a complex structure stabilized by multiple noncanonical interactions—revealed that a long-range tertiary interaction guides native RNA folding for both secondary and tertiary structure. This unanticipated nonhierarchical folding mechanism was directly validated by examining the consequences of concise disruption of the through-space interaction. Single-molecule, time-resolved RNA structure probing with TMO is poised to reveal a wide range of dynamic RNA folding processes and principles of RNA folding.
Citation
Time-Resolved, Single-Molecule, Correlated Chemical Probing of RNA
Jeffrey E. Ehrhardt and Kevin M. Weeks
Journal of the American Chemical Society 2020 142 (44), 18735-18740
DOI: 10.1021/jacs.0c06221