David Lawrence
Fred Eshelman Distinguished Professor, Jointly appointed with the UNC Schools of Pharmacy and Medicine
Not Accepting Doctoral Students
Kenan Laboratories A508919-962-8907
lawrencd@email.unc.edu
Group Website
Curriculum Vitae
Research Interests
Chemical Biology, Organic and Photochemistry, Molecular and Cell Biology
Research Synopsis
Living cells have been referred to as the test tubes of the 21st century. The creation of molecules that inhibit, probe, or alter the biochemistry of the cell lies at the nexus of chemistry and biology. The field of Chemical Biology seeks to correlate the underlying chemistry of life with the behavior of cells, tissues, and organisms. By revealing the nature of the molecular engine that drives cellular behavior Chemical Biology provides the molecular foundation upon which innovative therapies can be created for the entire spectrum of human afflictions.
The Lawrence research program is multifaceted, encompassing the fields of organic and peptide synthesis, photochemistry, enzymology, molecular and cell biology, and microscopy. This research team is engaged in the synthesis, characterization, and cell-based application of light-responsive agents, inhibitors, sensors, activators, proteins, and gene expression system, which are designed to manipulate and probe the biochemical pathways that control cell behavior. Disease states under investigation include cancer, neuropathologies, and inflammatory diseases.
Cellular Cyborgs
We have developed the cellular equivalent of cyborgs: Cells, erythrocytes, stem cells, containing implanted molecular devices, phototherapeutics. These tiny "terminators" are capable of launching multiple therapeutic agents at nearby diseased cells in response to wavelength-embedded commands. Since these light-triggered molecular launching pads detect red, far-red, and near IR deep-tissue penetrating light, they can receive external photo-instructions even when deeply embedded within tissue. In addition, the wavelength of light provoking drug launch can be pre-assigned, providing the ability to individually communicate with specific drugs in multi-drug carriers.
Angew. Chem. Intl. Ed. Engl., 2013, 52, 9936-9;
Angew. Chem. Intl. Ed. Engl., 2014, 53, 875-8;
Angew. Chem. Intl. Ed. Engl., 2014, 53, 10945 - 8.
Multicolor Monitoring of Enzyme Action
Conventional strategies for identifying the biochemical basis of tumorigenesis and metastasis rely upon the search for up-, or down-, regulated genes and proteins. However, the complexity and heterogeneity of many forms of cancer make it clear that this approach alone is not sufficient for extracting the information necessary to generate diagnostic and prognostic biomarkers. This biomedical imperative dictates the development of a series of new cellular and molecular strategies to tackle, what is admittedly, a devilishly difficult problem. We have developed an array of fluorescent sensors of protein remodeling enzymes, kinases, phosphatases, demininases, and proteases, that furnish robust readouts of catalytic activity, >100 fold, across the visible spectrum and into the near infrared. Furthermore, these sensors are photophysically distinct, enabling multiple enzymatic activities to be simultaneously monitored.
Angew. Chem. Intl. Ed. Engl., 2013, 52, 2323 - 5;
Angew. Chem. Intl. Ed. Engl., 2014, 53, 3975 - 8;
ACS Chem. Biol., 2015, 10, 433 - 40.
Light-Triggered Perturbation of Cell Behavior
We have employed a combination of organic photochemistry, organic and peptide synthesis, protein design, molecular biology, cell biology and microscopy to control and manipulate dynamic biological phenomena. Our molecular constructs have been used to identify the "steering wheel" of the cell during chemotaxis, with relevance to metastasis, probe intracellular enzymatic activity during the stages of cell division, reveal the mechanisms of gene transcription in single cells, and "command" cells to move or die upon exposure to a brief flash of light.
Science, 2004, 303, 743 - 6;
eLife, 2013, 2, E00750;
Angew. Chem. Intl. Ed. Engl., 2014, 53, 10904 - 7.
Predictive Personalized Pharmacology
The advent of effective pharmacologic inhibitors for clinical applications has created a critical need for assessing their efficacy in disease models and in patient samples. Biochemical analyses of aberrant signaling pathways are informative in terms of identifying the best treatment option and predicting therapeutic effectiveness in individual patients. One of the most compelling issues in preclinical and clinical drug discovery is the ability to accurately monitor drug action and patient responsiveness. In collaboration with Nancy Allbritton (Departments of Chemistry and Biomedical Engineering), and an array of clinician scientists on the UNC campus, we are developing chemical probes that can predict the optimal therapeutic agents for cancer patients.
PLoS One, 2012, 7, e48867;
Analytical Chemistry, 2013, 85, 6136 - 42;
Analytical Chemistry, 2014, 86, 4573 - 80.
Professional Background
UCLA Ph.D., 1982; NIH Postdoctoral Fellow, The Rockefeller University, 1982-85; Assistant, Associate, and Full Professor of Chemistry, SUNY Buffalo, 1985-95; Professor of Biochemistry, The Albert Einstein College of Medicine, 1996-2007; Professor of Chemistry, Chemical Biology & Medicinal Chemistry, School of Pharmacy, and Pharmacology, School of Medicine, Investigator, Lineberger Cancer Center, 2007–present; Chair, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, 2011–present
News & Publications
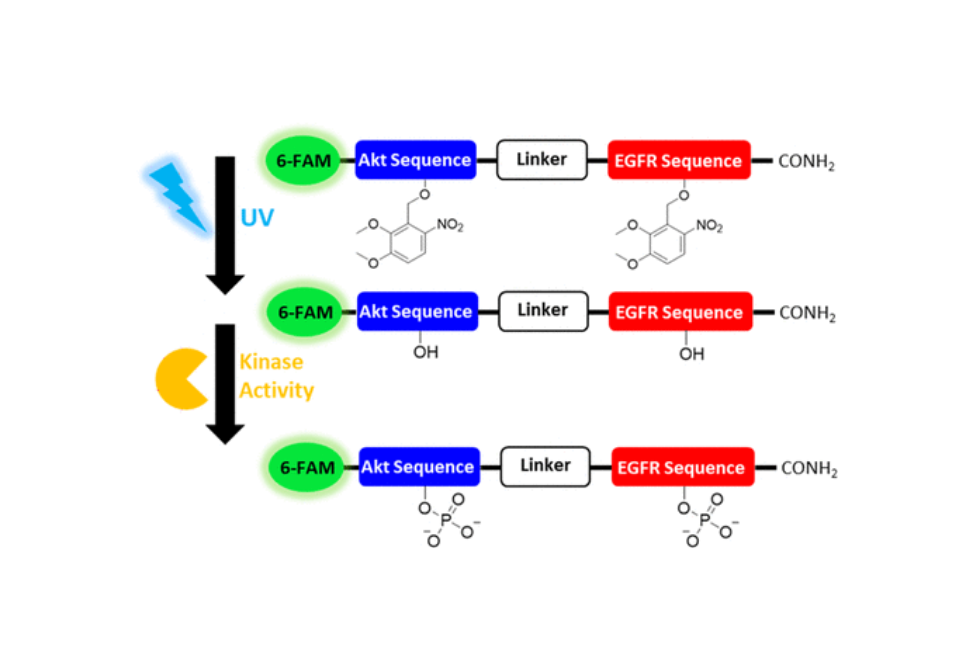
Peptide bioreporters were developed to perform multiplexed measurements of the activation of epidermal growth factor receptor kinase (EGFR), Akt kinase (Akt/protein kinase B), and proteases/peptidases in single cells.
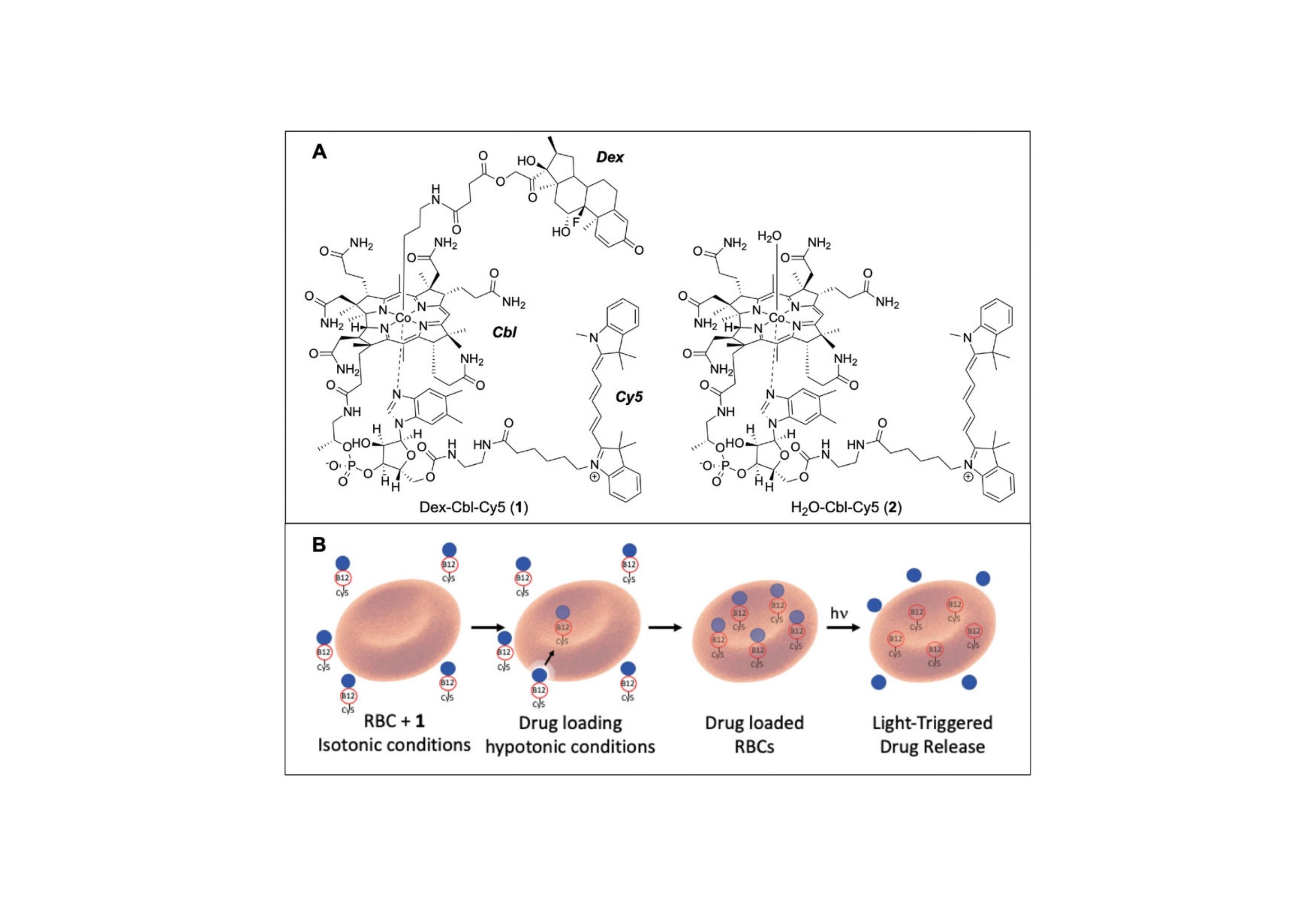
The design and application of a light-responsive red blood cell (RBC)-conveyed dexamethasone (Dex) construct that enables targeted drug delivery upon illumination of the inflamed site is described.